Python Plotting Basics#
Let’s take a look at some basic plotting with Python
If you’d like to follow along, connect to VDI then in a terminal run:
module use /g/data3/hh5/public/modules
module load conda/analysis3
jupyter notebook
This will start up a Jupyter Notebook on VDI, which is the same tool that this post is written in. Jupyter lets us work interactively with python and easily share code and plots.
# This is a python command
print("Hello world!")
# the output appears below
Hello world!
Sample data#
To start out, let’s get some sample data to work with. NCI has a data store with all of the Australian CMIP5 data. You can find this on NCI’s Data Catlogue
I like to use a library called Xarray to work with netCDF datasets. Let’s grab the monthly surface temperature from the ACCESS1-0 AMIP run and load it in
import xarray
datapath = "http://dapds00.nci.org.au/thredds/dodsC/rr3/CMIP5/output1/CSIRO-BOM/ACCESS1-0/amip/mon/atmos/Amon/r1i1p1/latest/tas/tas_Amon_ACCESS1-0_amip_r1i1p1_197901-200812.nc"
data = xarray.open_dataset(datapath)
I’m accessing the data remotely here using what’s called a THREDDS URL. You could just as easily use the /g/data filepath if you’re running on VDI.
To see what’s in the file we can just print it. This shows the dimensions, variables and metadata in the file:
print(data)
<xarray.Dataset>
Dimensions: (bnds: 2, lat: 145, lon: 192, time: 360)
Coordinates:
* time (time) datetime64[ns] 1979-01-16T12:00:00 1979-02-15 ...
* lat (lat) float64 -90.0 -88.75 -87.5 -86.25 -85.0 -83.75 -82.5 ...
* lon (lon) float64 0.0 1.875 3.75 5.625 7.5 9.375 11.25 13.12 15.0 ...
height float64 ...
Dimensions without coordinates: bnds
Data variables:
time_bnds (time, bnds) float64 ...
lat_bnds (lat, bnds) float64 ...
lon_bnds (lon, bnds) float64 ...
tas (time, lat, lon) float32 ...
Attributes:
institution: CSIRO (Commonwealth Scientific and Indus...
institute_id: CSIRO-BOM
experiment_id: amip
source: ACCESS1-0 2011. Atmosphere: AGCM v1.0 (N...
model_id: ACCESS1-0
forcing: GHG, Oz, SA, Sl, Vl, BC, OC, (GHG = CO2,...
parent_experiment_id: N/A
parent_experiment_rip: r1i1p1
branch_time: 0.0
contact: The ACCESS wiki: http://wiki.csiro.au/co...
history: CMIP5 compliant file produced from raw A...
references: See http://wiki.csiro.au/confluence/disp...
initialization_method: 1
physics_version: 1
tracking_id: 7cfe11fc-5b1c-457d-812b-e95f45e7def4
version_number: v20120115
product: output
experiment: AMIP
frequency: mon
creation_date: 2012-02-17T05:21:53Z
Conventions: CF-1.4
project_id: CMIP5
table_id: Table Amon (01 February 2012) 01388cb450...
title: ACCESS1-0 model output prepared for CMIP...
parent_experiment: N/A
modeling_realm: atmos
realization: 1
cmor_version: 2.8.0
DODS_EXTRA.Unlimited_Dimension: time
Accessing variables#
Variables are accessed using dot syntax. Just like the full dataset we can print individual variables to see their metadata:
surface_temp = data.tas
print(surface_temp)
<xarray.DataArray 'tas' (time: 360, lat: 145, lon: 192)>
[10022400 values with dtype=float32]
Coordinates:
* time (time) datetime64[ns] 1979-01-16T12:00:00 1979-02-15 ...
* lat (lat) float64 -90.0 -88.75 -87.5 -86.25 -85.0 -83.75 -82.5 ...
* lon (lon) float64 0.0 1.875 3.75 5.625 7.5 9.375 11.25 13.12 15.0 ...
height float64 ...
Attributes:
standard_name: air_temperature
long_name: Near-Surface Air Temperature
units: K
cell_methods: time: mean
cell_measures: area: areacella
history: 2012-02-17T05:21:51Z altered by CMOR: Treated scalar d...
associated_files: baseURL: http://cmip-pcmdi.llnl.gov/CMIP5/dataLocation...
Selecting data#
Before we can make a plot we first need to reduce the 3D data to 2D. One of the nice things about Xarray is that you can select data by a coordinate value, rather than manualy working out an index:
surface_temp_slice = surface_temp.sel(time = '1984-03')
print(surface_temp_slice.time)
<xarray.DataArray 'time' (time: 1)>
array(['1984-03-16T12:00:00.000000000'], dtype='datetime64[ns]')
Coordinates:
* time (time) datetime64[ns] 1984-03-16T12:00:00
height float64 ...
Attributes:
bounds: time_bnds
axis: T
long_name: time
standard_name: time
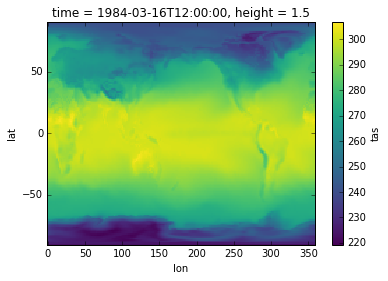
Plotting#
Now that we’ve selected our data we can get plotting
The notebook requires a special %matplotlib inline command to show plots, which isn’t needed if you’re writing a script.
To make a quick plot we can just call .plot() on a 2d variable:
%matplotlib inline
import matplotlib.pyplot as plt
surface_temp_slice.plot()
plt.show()